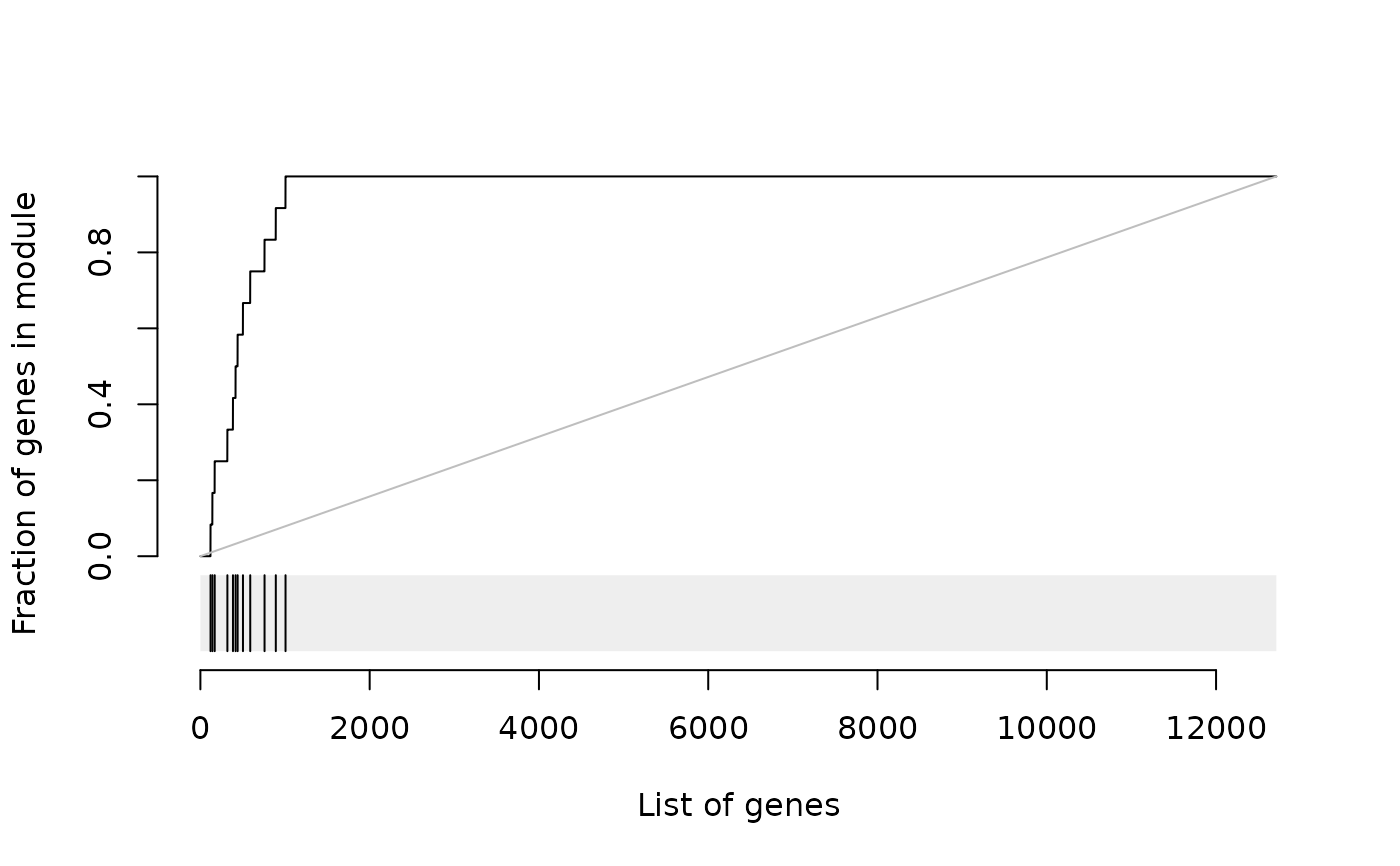
Create an evidence plot for a module
evidencePlot.RdCreate an evidence plot for a module
Usage
evidencePlot(
l,
m,
mset = "all",
rug = TRUE,
roc = TRUE,
filter = FALSE,
unique = TRUE,
add = FALSE,
col = "black",
col.rug = "#eeeeee",
gene.labels = NULL,
gene.colors = NULL,
gene.lines = 1,
gl.cex = 1,
style = "roc",
lwd = 1,
lty = 1,
rug.size = 0.2,
legend = NULL,
...
)Arguments
- l
sorted list of HGNC gene identifiers
- m
character vector of modules for which the plot should be created
- mset
Which module set to use (see tmodUtest for details)
- rug
if TRUE, draw a rug-plot beneath the ROC curve
- roc
if TRUE, draw a ROC curve above the rug-plot
- filter
if TRUE, genes not defined in the module set will be removed
- unique
if TRUE, duplicates will be removed
- add
if TRUE, the plot will be added to the existing plot
- col
a character vector color to be used
- col.rug
a character value specifying the color of the rug
- gene.labels
if TRUE, gene names are shown; alternatively, a named character vector with gene labels to be shown, or NULL (default) for no labels (option evaluated only if rug is plotted)
- gene.colors
NULL (default) or a character vectors indicating the color for each gene. Either a named vector or a vector with the same order of genes as `l`.
- gene.lines
a number or a vector of numbers; line width for marking the genes on the rug (default=1). If the vector is named, the names should be gene ids.
- gl.cex
Text cex (magnification) for gene labels
- style
"roc" for receiver-operator characteristic curve (default), and "gsea" for GSEA-style (Kaplan-Meier like plot)
- lwd
line width (see par())
- lty
line type (see par())
- rug.size
fraction of the plot that should show the rug. If rug.size is 0, rug is not drawn. If rug.size is 1, ROC curve is not drawn.
- legend
position of the legend. If NULL, no legend will be drawn
- ...
Further parameters passed to the plotting function
Details
This function creates an evidence plot for a module, based on an ordered list of genes. By default, the plot shows the receiving operator characteristic (ROC) curve and a rug below, which indicates the distribution of the module genes in the sorted list.
Several styles of the evidence plot are possible: * roc (default): a receiver-operator characteristic like curve; the area under the curve corresponds to the effect size (AUC) * roc_absolute: same as above, but the values are not scaled by the total number of genes in a module * gsea * enrichment: the curve shows relative enrichment at the given position